|
chr11_-_82674546
|
2.826
|
|
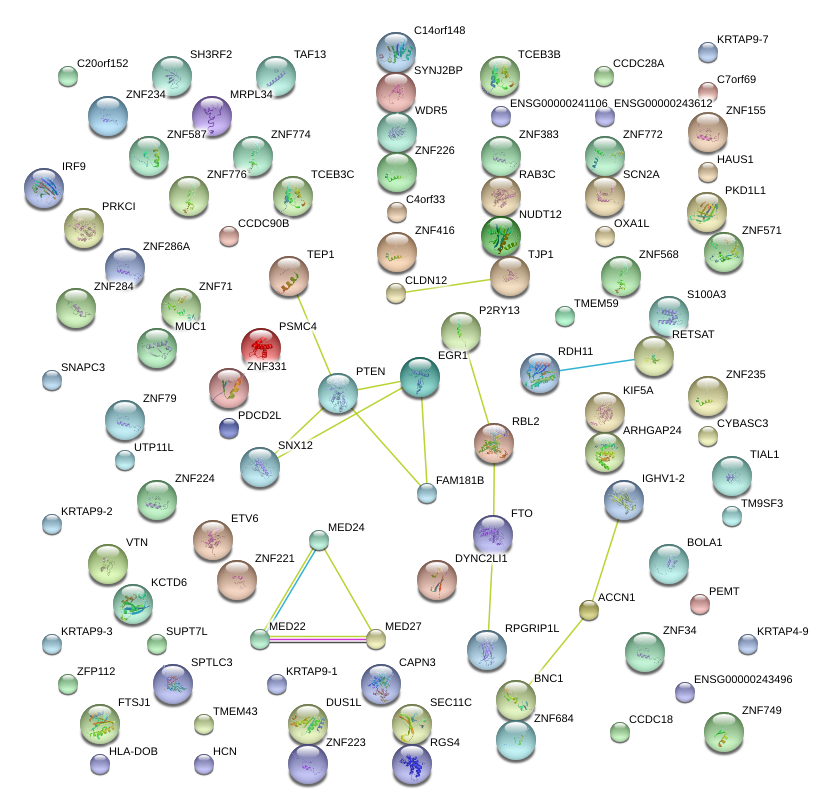
CCDC90B
|
coiled-coil domain containing 90B
|
|
chr11_-_82675024
|
1.949
|
NM_021825
|
CCDC90B
|
coiled-coil domain containing 90B
|
|
chr19_-_49309102
|
1.557
|
|
LOC100379224
|
hypothetical LOC100379224
|
|
chr19_+_49248003
|
1.431
|
NM_013361
|
ZNF223
|
zinc finger protein 223
|
|
chr19_+_49309350
|
1.390
|
NM_013362
|
ZNF225
|
zinc finger protein 225
|
|
chr19_+_49361054
|
1.260
|
NM_001146220
NM_001032373
NM_001032374
NM_015919
NM_001032372
|
ZNF226
|
zinc finger protein 226
|
|
chr19_-_62680703
|
1.259
|
NM_001024596
NM_001144068
|
ZNF772
|
zinc finger protein 772
|
|
chr19_+_49180171
|
1.023
|
NM_003445
NM_198089
|
ZNF155
|
zinc finger protein 155
|
|
chr9_+_129226473
|
0.960
|
NM_007135
|
ZNF79
|
zinc finger protein 79
|
|
chr9_+_129226535
|
0.949
|
|
ZNF79
|
zinc finger protein 79
|
|
chr17_+_36515166
|
0.924
|
NM_001146041
|
KRTAP4-9
|
keratin associated protein 4-9
|
|
chr19_+_49290319
|
0.894
|
NM_013398
|
ZNF224
|
zinc finger protein 224
|
|
chr4_+_86615275
|
0.880
|
NM_001025616
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr7_+_89870707
|
0.878
|
|
CLDN12
|
claudin 12
|
|
chr19_+_49147219
|
0.869
|
NM_013359
|
ZNF221
|
zinc finger protein 221
|
|
chr19_-_49552638
|
0.846
|
NM_001083335
NM_013380
|
ZFP112
|
zinc finger protein 112 homolog (mouse)
|
|
chr1_+_40769877
|
0.821
|
|
ZNF684
|
zinc finger protein 684
|
|
chr9_+_135994734
|
0.789
|
NM_052821
|
WDR5
|
WD repeat domain 5
|
|
chr9_-_133945064
|
0.766
|
NM_004269
|
CRSP8P
MED27
|
mediator complex subunit 27 pseudogene
mediator complex subunit 27
|
|
chr9_-_129226312
|
0.757
|
|
|
|
|
chr14_-_19951417
|
0.732
|
NM_007110
|
TEP1
|
telomerase-associated protein 1
|
|
chr2_+_43854681
|
0.689
|
NM_001193464
NM_015522
NM_016008
|
DYNC2LI1
|
dynein, cytoplasmic 2, light intermediate chain 1
|
|
chr14_-_67232172
|
0.685
|
NM_016026
|
RDH11
|
retinol dehydrogenase 11 (all-trans/9-cis/11-cis)
|
|
chr19_+_17277476
|
0.676
|
NM_023937
|
MRPL34
|
mitochondrial ribosomal protein L34
|
|
chr19_+_42400908
|
0.651
|
|
ZNF383
|
zinc finger protein 383
|
|
chr7_+_89870647
|
0.642
|
NM_001185072
NM_001185073
NM_012129
|
CLDN12
|
claudin 12
|
|
chr14_+_22305570
|
0.634
|
NM_005015
|
OXA1L
|
oxidase (cytochrome c) assembly 1-like
|
|
chr18_-_42815965
|
0.627
|
NM_016427
|
TCEB3B
|
transcription elongation factor B polypeptide 3B (elongin A2)
|
|
chr9_-_133944877
|
0.625
|
|
MED27
|
mediator complex subunit 27
|
|
chr19_+_42099073
|
0.571
|
NM_198539
|
ZNF568
|
zinc finger protein 568
|
|
chr20_+_12937616
|
0.568
|
NM_018327
|
SPTLC3
|
serine palmitoyltransferase, long chain base subunit 3
|
|
chr19_+_49337568
|
0.567
|
|
ZNF234
|
zinc finger protein 234
|
|
chr19_+_49337548
|
0.562
|
NM_001144824
NM_006630
|
ZNF234
|
zinc finger protein 234
|
|
chr17_-_18526296
|
0.555
|
NM_001145045
|
ZNF286B
|
zinc finger protein 286B
|
|
chr14_+_23700261
|
0.541
|
NM_006084
|
IRF9
|
interferon regulatory factor 9
|
|
chr1_+_38250953
|
0.534
|
NM_016037
|
UTP11L
|
UTP11-like, U3 small nucleolar ribonucleoprotein, (yeast)
|
|
chr5_+_145297490
|
0.533
|
|
SH3RF2
|
SH3 domain containing ring finger 2
|
|
chr11_+_65024889
|
0.532
|
|
MALAT1
|
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding)
|
|
chr10_-_121345968
|
0.527
|
|
TIAL1
|
TIA1 cytotoxic granule-associated RNA binding protein-like 1
|
|
chr3_+_58459131
|
0.518
|
NM_153331
|
KCTD6
|
potassium channel tetramerisation domain containing 6
|
|
chr8_-_145983457
|
0.517
|
NM_030580
|
ZNF34
|
zinc finger protein 34
|
|
chr1_+_40769819
|
0.504
|
NM_152373
|
ZNF684
|
zinc finger protein 684
|
|
chr16_+_52295545
|
0.492
|
|
FTO
|
fat mass and obesity associated
|
|
chr3_+_171422940
|
0.469
|
|
PRKCI
|
protein kinase C, iota
|
|
chr1_+_38250995
|
0.456
|
|
UTP11L
|
UTP11-like, U3 small nucleolar ribonucleoprotein, (yeast)
|
|
chr20_+_34019942
|
0.449
|
NM_080834
|
C20orf152
|
chromosome 20 open reading frame 152
|
|
chr19_-_49097774
|
0.445
|
|
LOC100505715
|
hypothetical LOC100505715
|
|
chr19_-_42098758
|
0.444
|
NM_001171979
|
ZNF829
|
zinc finger protein 829
|
|
chr2_-_85435074
|
0.438
|
NM_017750
|
RETSAT
|
retinol saturase (all-trans-retinol 13,14-reductase)
|
|
chr17_+_36636404
|
0.433
|
NM_031961
|
KRTAP9-2
|
keratin associated protein 9-2
|
|
chr19_-_62782066
|
0.423
|
|
ZNF416
|
zinc finger protein 416
|
|
chr17_+_15543771
|
0.410
|
|
ZNF286A
|
zinc finger protein 286A
|
|
chr19_+_61798434
|
0.406
|
NM_021216
|
ZNF71
|
zinc finger protein 71
|
|
chr1_+_161305188
|
0.401
|
NM_005613
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr7_+_47801413
|
0.398
|
NM_025031
|
PKD1L1
C7orf69
|
polycystic kidney disease 1 like 1
chromosome 7 open reading frame 69
|
|
chr1_+_54291847
|
0.397
|
NM_153035
|
TCEANC2
|
transcription elongation factor A (SII) N-terminal and central domain containing 2
|
|
chr19_+_49268106
|
0.384
|
NM_001037813
|
ZNF284
|
zinc finger protein 284
|
|
chr19_+_39587108
|
0.382
|
NM_032346
|
PDCD2L
|
programmed cell death 2-like
|
|
chr14_-_69953433
|
0.374
|
NM_018373
|
SYNJ2BP
|
synaptojanin 2 binding protein
|
|
chr19_+_62950001
|
0.373
|
|
ZNF776
|
zinc finger protein 776
|
|
chr17_-_17435631
|
0.359
|
NM_148172
|
PEMT
|
phosphatidylethanolamine N-methyltransferase
|
|
chr1_+_148137778
|
0.358
|
NM_016074
|
BOLA1
|
bolA homolog 1 (E. coli)
|
|
chr19_-_62781883
|
0.351
|
NM_017879
|
ZNF416
|
zinc finger protein 416
|
|
chr14_-_76958984
|
0.341
|
NM_138791
NM_001113475
|
C14orf148
|
chromosome 14 open reading frame 148
|
|
chr1_+_93418866
|
0.334
|
|
CCDC18
|
coiled-coil domain containing 18
|
|
chr17_+_36599664
|
0.324
|
NM_001190460
|
KRTAP9-1
|
keratin associated protein 9-1
|
|
chr15_-_81744469
|
0.320
|
NM_001717
|
BNC1
|
basonuclin 1
|
|
chr19_-_42098919
|
0.318
|
NM_001037232
|
ZNF829
|
zinc finger protein 829
|
|
chr1_+_161305558
|
0.316
|
NM_001113381
|
RGS4
|
regulator of G-protein signaling 4
|
|
chr4_+_130236731
|
0.313
|
NM_001099783
|
C4orf33
|
chromosome 4 open reading frame 33
|
|
chr10_-_98315090
|
0.310
|
|
TM9SF3
|
transmembrane 9 superfamily member 3
|
|
chr11_-_60880799
|
0.303
|
NM_001161452
|
CYBASC3
|
cytochrome b, ascorbate dependent 3
|
|
chr19_-_49500966
|
0.301
|
NM_004234
|
ZNF235
|
zinc finger protein 235
|
|
chr1_-_54291634
|
0.297
|
NM_004872
|
TMEM59
|
transmembrane protein 59
|
|
chr1_-_206125877
|
0.295
|
|
|
|
|
chr1_-_109420091
|
0.294
|
NM_005645
|
TAF13
|
TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa
|
|
chr17_-_77616803
|
0.293
|
|
DUS1L
|
dihydrouridine synthase 1-like (S. cerevisiae)
|
|
chr15_+_40427592
|
0.292
|
NM_212465
|
CAPN3
|
calpain 3, (p94)
|
|
chr1_-_151788341
|
0.286
|
NM_002960
|
S100A3
|
S100 calcium binding protein A3
|
|
chr18_+_54958095
|
0.284
|
NM_033280
|
SEC11C
|
SEC11 homolog C (S. cerevisiae)
|
|
chr19_+_58715988
|
0.284
|
NM_018555
|
ZNF331
|
zinc finger protein 331
|
|
chr17_-_77616968
|
0.282
|
NM_022156
|
DUS1L
|
dihydrouridine synthase 1-like (S. cerevisiae)
|
|
chr19_+_62638474
|
0.280
|
NM_001023561
|
ZNF749
|
zinc finger protein 749
|
|
chr17_+_15543615
|
0.277
|
NM_001130842
NM_020652
|
ZNF286A
|
zinc finger protein 286A
|
|
chr16_+_52025874
|
0.271
|
|
RBL2
|
retinoblastoma-like 2 (p130)
|
|
chr9_+_15412875
|
0.269
|
|
SNAPC3
|
small nuclear RNA activating complex, polypeptide 3, 50kDa
|
|
chr2_+_165858586
|
0.267
|
NM_021007
|
SCN2A
|
sodium channel, voltage-gated, type II, alpha subunit
|
|
chr6_+_139136349
|
0.264
|
NM_015439
|
CCDC28A
|
coiled-coil domain containing 28A
|
|
chr5_-_102926349
|
0.260
|
NM_031438
|
NUDT12
|
nudix (nucleoside diphosphate linked moiety X)-type motif 12
|
|
chrX_+_48219344
|
0.259
|
NM_012280
NM_177439
|
FTSJ1
|
FtsJ homolog 1 (E. coli)
|
|
chr16_-_52295214
|
0.258
|
NM_001127897
NM_015272
|
RPGRIP1L
|
RPGRIP1-like
|
|
chr18_+_41938295
|
0.258
|
NM_138443
|
HAUS1
|
HAUS augmin-like complex, subunit 1
|
|
chr6_-_32892705
|
0.258
|
NM_002120
|
HLA-DOB
|
major histocompatibility complex, class II, DO beta
|
|
chr15_+_88696480
|
0.257
|
NM_001004309
|
ZNF774
|
zinc finger protein 774
|
|
chr3_-_152529999
|
0.254
|
NM_176894
|
P2RY13
|
purinergic receptor P2Y, G-protein coupled, 13
|
|
chr17_-_23721430
|
0.250
|
|
VTN
|
vitronectin
|
|
chr17_-_35464137
|
0.250
|
|
MED24
|
mediator complex subunit 24
|
|
chr19_+_45168872
|
0.250
|
NM_006503
NM_153001
|
PSMC4
|
proteasome (prosome, macropain) 26S subunit, ATPase, 4
|
|
chr3_+_14141633
|
0.249
|
|
TMEM43
|
transmembrane protein 43
|
|
chr2_-_27739947
|
0.247
|
NM_014860
|
SUPT7L
|
suppressor of Ty 7 (S. cerevisiae)-like
|
|
chr19_+_17239264
|
0.247
|
|
BABAM1
|
BRISC and BRCA1 A complex member 1
|
|
chr5_+_57914658
|
0.244
|
NM_138453
|
RAB3C
|
RAB3C, member RAS oncogene family
|
|
chr3_+_14141583
|
0.243
|
|
TMEM43
|
transmembrane protein 43
|
|
chr9_-_114814286
|
0.243
|
NM_001101338
|
ZNF883
|
zinc finger protein 883
|
|
chr17_-_23721351
|
0.240
|
|
VTN
|
vitronectin
|
|
chr15_-_27901628
|
0.239
|
|
TJP1
|
tight junction protein 1 (zona occludens 1)
|
|
chr12_+_11694172
|
0.237
|
|
ETV6
|
ets variant 6
|
|
chr17_-_23721485
|
0.237
|
NM_000638
|
VTN
|
vitronectin
|
|
chr15_+_20384835
|
0.232
|
NM_001102610
NM_052903
|
TUBGCP5
|
tubulin, gamma complex associated protein 5
|
|
chr16_+_52025863
|
0.231
|
|
RBL2
|
retinoblastoma-like 2 (p130)
|
|
chr6_-_30766621
|
0.226
|
NM_007243
|
NRM
|
nurim (nuclear envelope membrane protein)
|
|
chr11_-_58368555
|
0.226
|
NM_145016
|
GLYATL2
|
glycine-N-acyltransferase-like 2
|
|
chr17_-_23721468
|
0.226
|
|
VTN
|
vitronectin
|
|
chr18_-_23019176
|
0.226
|
NM_031422
|
CHST9
|
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 9
|
|
chr17_-_77616953
|
0.222
|
|
DUS1L
|
dihydrouridine synthase 1-like (S. cerevisiae)
|
|
chr16_+_52025891
|
0.218
|
|
RBL2
|
retinoblastoma-like 2 (p130)
|
|
chr19_+_45194782
|
0.218
|
NM_178544
|
ZNF546
|
zinc finger protein 546
|
|
chr2_+_95195260
|
0.213
|
|
ZNF2
|
zinc finger protein 2
|
|
chr17_-_23721420
|
0.213
|
|
VTN
|
vitronectin
|
|
chr19_+_49455872
|
0.212
|
NM_181756
|
ZNF233
|
zinc finger protein 233
|
|
chr11_+_67130898
|
0.210
|
NM_001166102
NM_007103
|
NDUFV1
|
NADH dehydrogenase (ubiquinone) flavoprotein 1, 51kDa
|
|
chr15_+_39853923
|
0.208
|
NM_001128608
NM_014994
|
MAPKBP1
|
mitogen-activated protein kinase binding protein 1
|
|
chr11_+_36572615
|
0.208
|
NM_138787
|
C11orf74
|
chromosome 11 open reading frame 74
|
|
chr11_+_72658586
|
0.208
|
NM_176798
|
P2RY6
|
pyrimidinergic receptor P2Y, G-protein coupled, 6
|
|
chrX_+_105932536
|
0.205
|
NM_017752
NM_198881
|
TBC1D8B
|
TBC1 domain family, member 8B (with GRAM domain)
|
|
chr18_-_41938196
|
0.205
|
NM_001001937
|
ATP5A1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1, cardiac muscle
|
|
chr19_-_3752809
|
0.199
|
NM_002378
|
MATK
|
megakaryocyte-associated tyrosine kinase
|
|
chr5_+_178219519
|
0.198
|
NM_058230
|
ZNF354B
|
zinc finger protein 354B
|
|
chr19_+_62885148
|
0.198
|
NM_138347
|
ZNF551
|
zinc finger protein 551
|
|
chr10_-_131799051
|
0.196
|
|
LOC387723
|
hypothetical LOC387723
|
|
chr19_+_62949972
|
0.194
|
NM_173632
|
ZNF776
|
zinc finger protein 776
|
|
chr5_+_14493905
|
0.194
|
|
TRIO
|
triple functional domain (PTPRF interacting)
|
|
chr6_+_35812836
|
0.192
|
NM_145028
|
C6orf81
|
chromosome 6 open reading frame 81
|
|
chr1_-_33275051
|
0.192
|
|
AK2
|
adenylate kinase 2
|
|
chr21_-_35174742
|
0.191
|
|
RUNX1
|
runt-related transcription factor 1
|
|
chr17_-_44847215
|
0.190
|
NM_002634
|
PHB
|
prohibitin
|
|
chr7_+_98940537
|
0.189
|
|
ZKSCAN5
|
zinc finger with KRAB and SCAN domains 5
|
|
chr3_+_14141544
|
0.189
|
|
TMEM43
|
transmembrane protein 43
|
|
chrX_+_24621984
|
0.188
|
NM_016937
|
POLA1
|
polymerase (DNA directed), alpha 1, catalytic subunit
|
|
chr19_-_63205948
|
0.188
|
|
ZNF606
|
zinc finger protein 606
|
|
chr2_+_218843358
|
0.188
|
NM_001077399
NM_015488
|
PNKD
|
paroxysmal nonkinesigenic dyskinesia
|
|
chr19_-_49131250
|
0.188
|
NM_003425
|
ZNF45
|
zinc finger protein 45
|
|
chr19_-_63150825
|
0.188
|
NM_005773
|
ZNF256
|
zinc finger protein 256
|
|
chr16_+_52025845
|
0.187
|
NM_005611
|
RBL2
|
retinoblastoma-like 2 (p130)
|
|
chr1_-_33275039
|
0.185
|
|
AK2
|
adenylate kinase 2
|
|
chr19_+_17239148
|
0.184
|
NM_001033549
NM_014173
|
BABAM1
|
BRISC and BRCA1 A complex member 1
|
|
chr5_-_111120817
|
0.181
|
NM_004772
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr8_-_56848423
|
0.180
|
NM_152417
|
TMEM68
|
transmembrane protein 68
|
|
chr3_+_14141553
|
0.179
|
|
TMEM43
|
transmembrane protein 43
|
|
chr19_+_51804260
|
0.179
|
|
CALM3
|
calmodulin 3 (phosphorylase kinase, delta)
|
|
chr7_+_128290162
|
0.178
|
|
ATP6V1F
|
ATPase, H+ transporting, lysosomal 14kDa, V1 subunit F
|
|
chr3_+_125586241
|
0.178
|
|
KALRN
|
kalirin, RhoGEF kinase
|
|
chr12_-_102413875
|
0.177
|
NM_198521
NM_001099336
|
C12orf42
|
chromosome 12 open reading frame 42
|
|
chr9_+_70161634
|
0.174
|
NM_021965
|
PGM5
|
phosphoglucomutase 5
|
|
chr12_-_47735373
|
0.173
|
NM_003482
|
MLL2
|
myeloid/lymphoid or mixed-lineage leukemia 2
|
|
chr3_+_171422908
|
0.172
|
NM_002740
|
PRKCI
|
protein kinase C, iota
|
|
chr1_-_33275015
|
0.172
|
|
AK2
|
adenylate kinase 2
|
|
chr16_-_20246309
|
0.171
|
NM_001007240
NM_001007241
NM_001007242
NM_001502
|
GP2
|
glycoprotein 2 (zymogen granule membrane)
|
|
chr11_-_133628348
|
0.163
|
NM_001037304
NM_001037305
NM_014174
NM_199297
NM_199298
|
THYN1
|
thymocyte nuclear protein 1
|
|
chr12_+_119224506
|
0.163
|
NM_012240
|
SIRT4
|
sirtuin 4
|
|
chr16_+_68542126
|
0.160
|
NM_001136214
|
CLEC18A
|
C-type lectin domain family 18, member A
|
|
chr19_+_49221333
|
0.159
|
NM_001129996
NM_013360
|
ZNF284
ZNF222
|
zinc finger protein 284
zinc finger protein 222
|
|
chr5_-_95184221
|
0.157
|
NM_001118890
NM_002064
|
GLRX
|
glutaredoxin (thioltransferase)
|
|
chr8_-_109330116
|
0.152
|
NM_001568
|
EIF3E
|
eukaryotic translation initiation factor 3, subunit E
|
|
chr1_-_3763620
|
0.150
|
NM_014704
|
KIAA0562
|
KIAA0562
|
|
chr17_+_36665161
|
0.150
|
NM_030975
|
KRTAP9-9
|
keratin associated protein 9-9
|
|
chr1_+_109827975
|
0.149
|
NM_153340
|
ATXN7L2
|
ataxin 7-like 2
|
|
chr2_-_230792844
|
0.148
|
NM_004509
NM_004510
NM_080424
|
SP110
|
SP110 nuclear body protein
|
|
chrX_+_77041612
|
0.148
|
NM_001866
|
COX7B
|
cytochrome c oxidase subunit VIIb
|
|
chr8_-_144770764
|
0.146
|
|
TSTA3
|
tissue specific transplantation antigen P35B
|
|
chr19_+_62972861
|
0.145
|
|
ZNF586
|
zinc finger protein 586
|
|
chr19_+_13090101
|
0.144
|
NM_052876
|
NACC1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing
|
|
chr2_+_63669596
|
0.142
|
NM_005917
|
MDH1
|
malate dehydrogenase 1, NAD (soluble)
|
|
chr17_-_3740785
|
0.141
|
NM_172206
|
CAMKK1
|
calcium/calmodulin-dependent protein kinase kinase 1, alpha
|
|
chr8_-_109330101
|
0.141
|
|
EIF3E
|
eukaryotic translation initiation factor 3, subunit E
|
|
chr1_-_145608965
|
0.140
|
NM_016361
|
ACP6
|
acid phosphatase 6, lysophosphatidic
|
|
chr4_+_109763119
|
0.138
|
|
RPL34
|
ribosomal protein L34
|
|
chr5_-_145542428
|
0.136
|
NM_020117
|
LARS
|
leucyl-tRNA synthetase
|
|
chr19_+_62554445
|
0.135
|
NM_020657
|
ZNF304
|
zinc finger protein 304
|
|
chr17_+_77910411
|
0.134
|
NM_207459
|
TEX19
|
testis expressed 19
|
|
chr3_+_199161446
|
0.133
|
NM_000996
|
RPL35A
|
ribosomal protein L35a
|
|
chr11_-_72182291
|
0.131
|
NM_006645
|
STARD10
|
StAR-related lipid transfer (START) domain containing 10
|
|
chr14_+_23175383
|
0.131
|
NM_005794
NM_182908
|
DHRS2
|
dehydrogenase/reductase (SDR family) member 2
|
|
chr1_-_25431544
|
0.129
|
NM_015484
NM_207170
|
SYF2
|
SYF2 homolog, RNA splicing factor (S. cerevisiae)
|
|
chr15_+_39853982
|
0.129
|
|
MAPKBP1
|
mitogen-activated protein kinase binding protein 1
|
|
chr3_+_171422917
|
0.128
|
|
PRKCI
|
protein kinase C, iota
|
|
chr17_-_44847214
|
0.127
|
|
PHB
|
prohibitin
|
|
chr2_+_95194903
|
0.121
|
NM_001017396
NM_021088
|
ZNF2
|
zinc finger protein 2
|
|
chr19_-_49597594
|
0.119
|
NM_152354
|
ZNF285
ZFP112
|
zinc finger protein 285
zinc finger protein 112 homolog (mouse)
|
|
chr1_-_11041490
|
0.117
|
|
SRM
|
spermidine synthase
|
|
chr1_+_3763704
|
0.117
|
NM_004402
|
DFFB
|
DNA fragmentation factor, 40kDa, beta polypeptide (caspase-activated DNase)
|
|
chr2_-_84540096
|
0.115
|
NM_003849
|
SUCLG1
|
succinate-CoA ligase, alpha subunit
|
|
chr2_+_27740254
|
0.114
|
|
SLC4A1AP
|
solute carrier family 4 (anion exchanger), member 1, adaptor protein
|
|
chr19_-_52045890
|
0.114
|
NM_004069
NM_021575
|
AP2S1
|
adaptor-related protein complex 2, sigma 1 subunit
|
|
chr19_+_49408523
|
0.114
|
NM_182490
|
ZNF227
|
zinc finger protein 227
|
|
chr8_+_27687821
|
0.112
|
NM_001017420
|
ESCO2
|
establishment of cohesion 1 homolog 2 (S. cerevisiae)
|
|
chr11_-_100959868
|
0.111
|
NM_004621
|
TRPC6
|
transient receptor potential cation channel, subfamily C, member 6
|
|
chr10_-_38305458
|
0.111
|
NM_145011
|
ZNF25
|
zinc finger protein 25
|
|
chr3_+_14141440
|
0.110
|
NM_024334
|
TMEM43
|
transmembrane protein 43
|
|
chr1_+_17571277
|
0.109
|
NM_207421
|
PADI6
|
peptidyl arginine deiminase, type VI
|